Variant Report
OpenCRAVAT provides a whole-page variant report that describes an individual variant. Try these examples below, which will open an OpenCRAVAT Variant Report page from the OpenCRAVAT public server.
OpenCRAVAT Variant Page example 1
OpenCRAVAT Variant Page example 2
OpenCRAVAT Variant Page example 3
OpenCRAVAT Variant Page example 4
To enable OpenCRAVAT Variant Report pages in your own installation of OpenCRAVAT, there are necessary annotation modules. Run the command below to install the required modules.
oc module install lollipop hgvs chasmplus civic cosmic cgc cgl target gnomad thousandgenomes hgdp esp6500 abraom uk10k_cohort pharmgkb clinvar clingen denovo gwas_catalog ess_gene gnomad_gene prec phi ghis loftool interpro gtex rvis ncbigene vest mutpred1 mutation_assessor fathmm phdsnpg phastcons phylop linsight ncrna pseudogene biogrid intact ndex mupit
The OpenCRAVAT Variant Report page can be launched with the following URL scheme:
http(s)://<OpenCRAVAT GUI domain>/result/nocache/variant.html?chrom=<chromosome>&pos=<genomic position>&ref_base=<reference base(s)>&alt_base=<alternate base(s)>
For example,
http://localhost:8060/result/nocache/variant.html?chrom=chr1&pos=12777320&ref_base=C&alt_base=T
If you open this URL the first time after starting oc gui, the page
will take some time to load since the gene mapper and several required
annotators need to be loaded.
The Variant Report page has the following sections.
Variant Annotations

single variant report variant annotations
This section includes basic information on the input variant, the mapping of the variant onto genes with variant consequences, mappings to UniProt, HGVS notation of variant consequences, and a protein diagram showing the location of the variant as well as Pfam and UniProt domain annotations.
Cancer
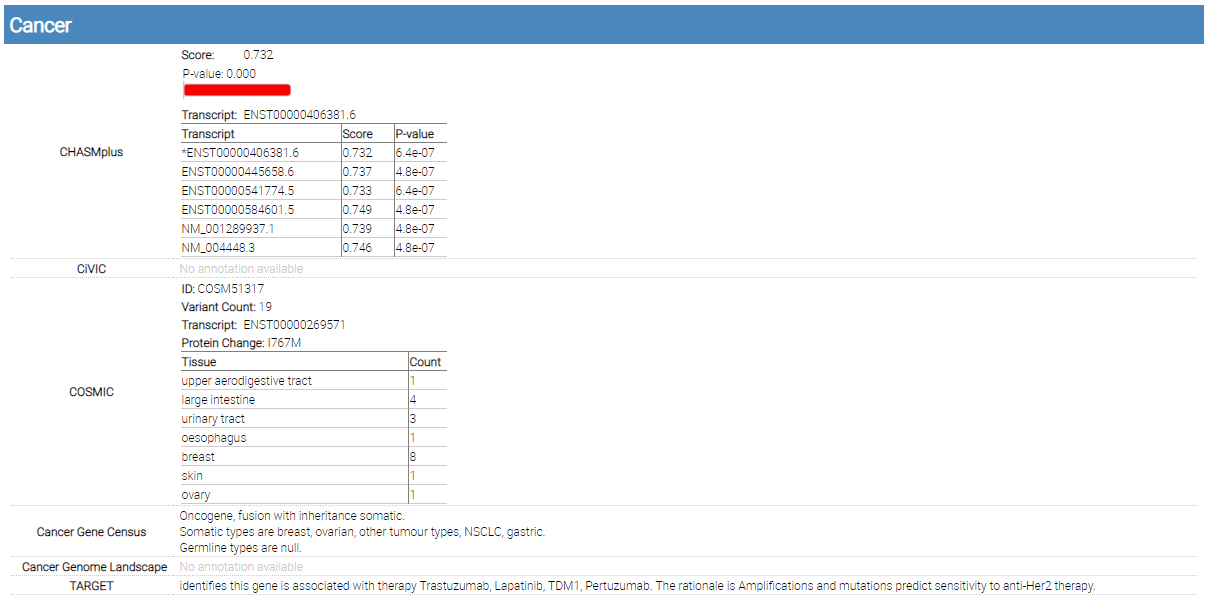
single variant report cancer
Several cancer-related annotations are included in this section: CHASMplus, CiVIC, COSMIC, Cancer Gene Census, Cancer Genome Landscape, and TARGET.
Population Allele Frequencies
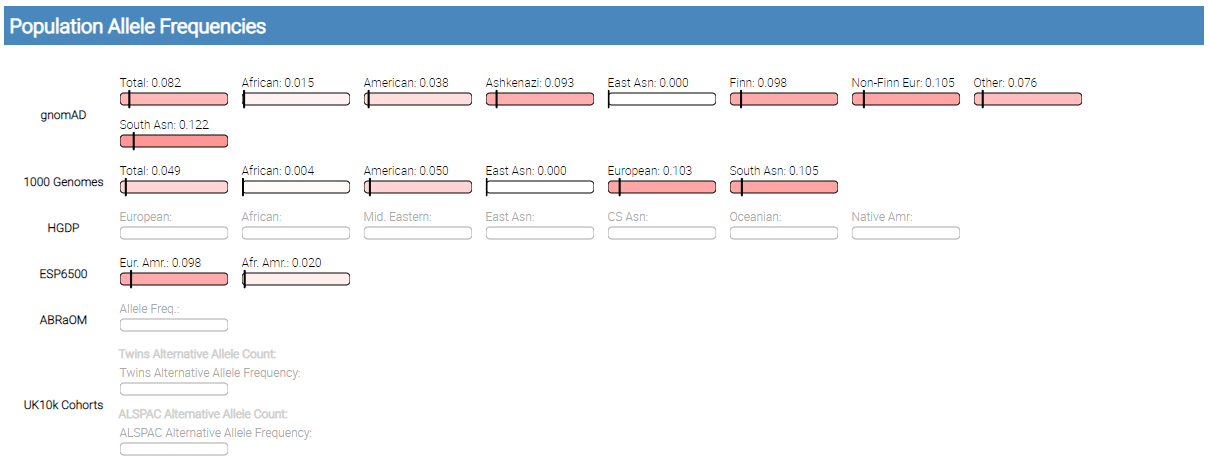
single variant report population allele frequencies
gnomAD, 1000 Genomes, HGDP, ESP6500, ABRaOM, and UK10K Cohorts allele frequency annotations are shown in this section.
Clinical Relevance
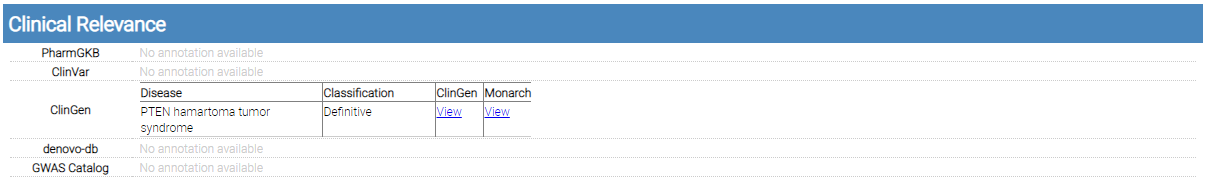
single variant report clinical relevance
PharmGKB, ClinVar, ClinGen, denovo-db, and GWAS Catalog annotations are shown in this section.
Gene

single variant report gene
Gene-level annotations are shown in this section: gnomAD Gene, P(rec), P(HI), GHIS, LoFtool, RVIS, and NCBI Gene.
Pathogenicity Prediction

single variant report pathogenicity prediction
Several variant effect prediction annotations are included in this section: VEST, PhDSNPg, MutPred, Mutation Assessor, and FATHMM.
Evolutionary Constraint

single variant report evolutionary constraint
PhastCons and PhyloP annotations are shown in this section.
Noncoding and Genomic Elements
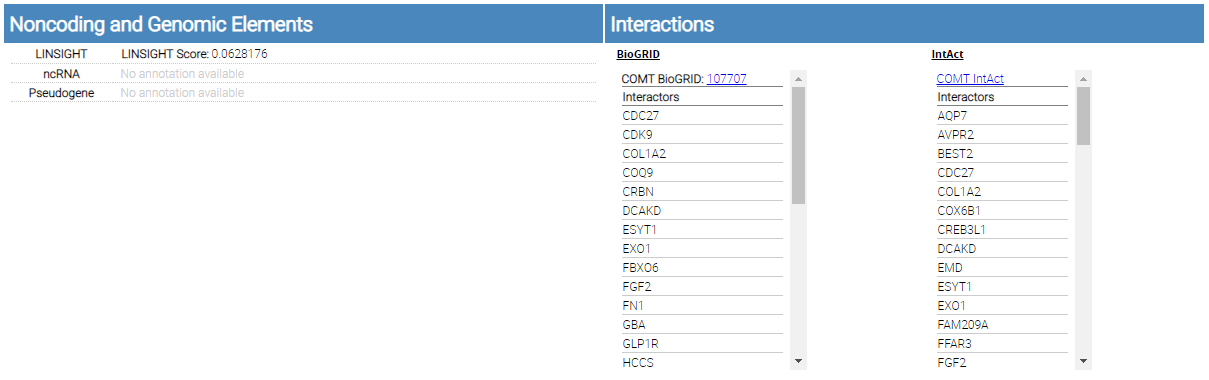
single variant report noncoding and genomic elements
This section includes annotations from LINSIGHT, ncRNA, Pseudogene, BioGRID, and IntAct.
Visualization

single variant report visualization
NDEx and MuPIT visualizations are shown in this section. In the NDEx visualization, use the dropdown menu to select different networks which include the gene impacted by the variant. The variant’s gene is shown in red. In MuPIT visualization, click the triangle at the left bottom and then click “Result” button to select different 3D protein structures to which the variant was mapped. Also, click the triangle at the top right to highlight TCGA mutations and UniProt-derived protein domains on the shown 3D structure.